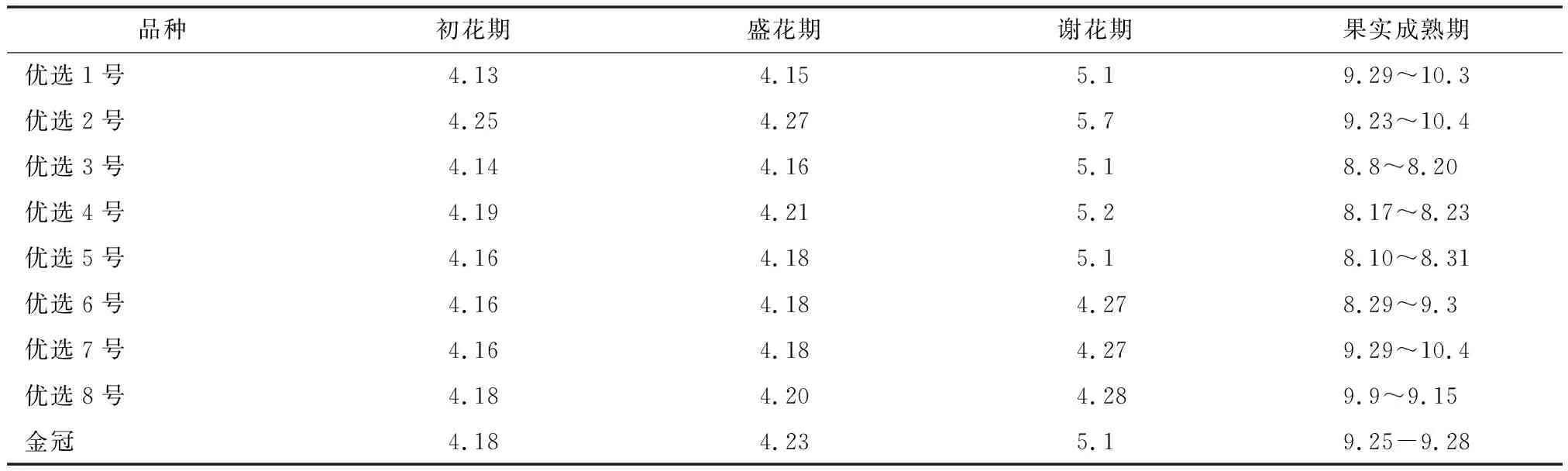
摘 要:【目的】研究不同抗性遗传背景棉蚜对氟啶虫胺腈和啶虫脒抗性机制差异。
【方法】利用Illumina高通量测序技术,分别对2个不同抗性遗传背景棉蚜(莎车县和精河县)的田间初始品系、啶虫脒抗性品系和氟啶虫胺腈抗性品系进行转录组测序,利用生物信息学方法比较分析2个不同抗性遗传背景棉蚜种群各品系差异表达基因。
【结果】莎车县氟啶虫胺腈抗性品系和啶虫脒抗性品系分别有806个和149个基因差异表达;精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系与精河县的田间初始品系相比分别有233个和160个基因差异表达。在莎车县氟啶虫胺腈抗性品系和啶虫脒抗性品系中,CYP6CY59、CYP6DC1和CYP380C45均上调表达,CYP6CY12和CYP380C46均下调表达;在精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系中,CYP380C46均上调表达,CYP6DC1均下调表达。此外,CYP380C45在莎车县氟啶虫胺腈抗性品系、精河县氟啶虫胺腈抗性品系和莎车县啶虫脒抗性品系中均上调表达;CYP6DC1在莎车县2个抗性品系中上调表达,但在精河县2个抗性品系中下调表达;相反,CYP380C46在精河县2个抗性品系中上调表达,但在莎车县2个抗性品系中下调表达。
【结论】有多个P450基因参与棉蚜对氟啶虫胺腈和啶虫脒的抗性。相同抗性遗传背景的棉蚜氟啶虫胺腈与啶虫脒抗性品系之间差异表达的P450基因存在差异,而且不同抗性遗传背景棉蚜的氟啶虫胺腈抗性品系之间以及啶虫脒抗性品系之间差异表达的P450基因也存在差异。
关键词:棉蚜;抗性;差异表达基因
中图分类号:S41"" 文献标志码:A"" 文章编号:1001-4330(2024)12-3078-11
0 引 言
【研究意义】棉蚜Aphis gossypii是棉花重要害虫之一[1,2]。化学防治不当可导致棉蚜对有机磷类、氨基甲酸酯类、拟除虫菊酯类和新烟碱类杀虫剂产生抗性[3]。由于新疆各棉区生态环境不同、棉花田间管理差异较大,不同地域棉蚜对新烟碱类杀虫剂抗性差异明显。因此,研究不同抗性遗传背景棉蚜抗药性机制差异性,对新疆棉蚜的合理防治及抗药性治理有重要意义。【前人研究进展】解毒代谢能力增强和靶标敏感性下降是棉蚜抗新烟碱类杀虫剂两个主要机制[4]。涉及棉蚜对新烟碱类杀虫剂解毒代谢的酶和蛋白主要包括细胞色素P450多功能氧化酶(Cytochrome P450 monooxygenases,P450)、UDP-葡糖基转移酶(Uridine diphosphate glycosyltransferases,UGT)和ABC转运蛋白(ATP-binding cassette transporters)。以往研究已证实P450基因、UGT基因和ABC转运蛋白基因的过量表达是棉蚜对新烟碱类杀虫剂产生抗性的重要原因[5-7]。另外,烟碱型乙酰胆碱受体(nicotinic acetylcholine receptor,nAChR)是新烟碱类杀虫剂的作用靶标[8],nAChR靶标位点突变是棉蚜对新烟碱类杀虫剂产生抗性的重要机制[9]。棉蚜nAChR β1亚基环D区域发生R81T、L80S、K264E和V62I突变涉及棉蚜对吡虫啉抗性[10-13],而且棉蚜nAChR β1、α1、α4-1、α4-2、α5和α7亚基表达显著下降也会导致棉蚜对新烟碱类杀虫剂抗性上升[14]。【本研究切入点】氟啶虫胺腈是一种砜亚胺类杀虫剂,被抗杀虫剂行动委员会(Insecticide Resistance Action Committee)归为4C类[15]。氟啶虫胺腈作为一种重要的替代杀虫剂被广泛用于防治粉虱类、飞虱类、蚜虫类等刺吸式害虫[16]。尽管氟啶虫胺腈是一种nAChR竞争性调节剂[16],但其与解毒代谢酶和nAChR相互作用方式不同于啶虫脒和吡虫啉等新烟碱类杀虫剂(4A 类)[15,17]。棉蚜地理种群差异而形成抗药性遗传背景不同可能影响其对杀虫剂抗性机制,而且棉蚜对不同杀虫剂抗性机制可能也存在差异。需选取不同抗性遗传背景棉蚜的氟啶虫胺腈和啶虫脒抗性品系进行转录组测序及差异基因比较,探明不同抗性遗传背景棉蚜对氟啶虫胺腈和啶虫脒的抗性机制差异。【拟解决的关键问题】研究利用Illumina高通量测序技术,分别对2个不同抗性遗传背景棉蚜(莎车县和精河县)的田间初始品系、啶虫脒抗性品系和氟啶虫胺腈抗性品系进行转录组测序,利用生物信息学方法比较分析2个不同抗性遗传背景棉蚜种群各品系差异表达基因,研究不同抗性遗传背景棉蚜对啶虫脒和氟啶虫胺腈抗性机制差异,为棉蚜抗药性治理提供理论支撑。
1 材料与方法
1.1 材 料
2个棉蚜田间品系于2019年7月分别采自新疆莎车县(77°281′E,38°546′N)和精河县(82°896′E,44°588′N)棉田,在中国农业科学院廊坊科研中试基地温室用棉花幼苗饲养,饲养条件为温度(26 ± 5)℃,70%±10% RH,光周期16∶8 L/D。棉花品种为中棉49,由中国农业科学院棉花所提供。每个田间种群分别被分为3个品系,第1个品系被连续暴露于氟啶虫胺腈24代,作为氟啶虫胺腈抗性品系;第2个品系被连续暴露于抗啶虫脒24代,作为啶虫脒抗性品系;第3个品系不接触任何杀虫剂,作为田间初始品系进行饲养,共6个品系,即:莎车县田间初始品系(Yarkant-FS)、莎车县氟啶虫胺腈抗性品系(Yarkant-SulR)、莎车县啶虫脒抗性品系(Yarkant-AceR)、精河县田间初始品系(Jinghe-FS)、精河县氟啶虫胺腈抗性品系(Jinghe-SulR)、精河县啶虫脒抗性品系(Jinghe-AceR)。
1.2 方 法
1.2.1 RNA提取及测序
在莎车县和精河县的田间初始品系、氟啶虫胺腈抗性品系和啶虫脒抗性品系中,分别随机选取体型一致的无翅成蚜40头,分装于1.5 mL RNase-free 离心管,迅速液氮处理,并置于-80℃低温冰箱保存。每个品系3个重复,共计6品系18个样品。采用Trizol法提取样品RNA,通过NanoDrop 2000超微量分光光度计检测RNA纯度及浓度,利用Agient 2100生物分析仪检测RNA完整性,采用Illumina NovaSeq6000测序平台进行转录组测序。转录组测序委托北京百迈客生物科技有限公司完成。
1.2.2 测序数据评估、组装及功能注释
通过去除原始数据中含有接头的和低质量的Reads(N比例大于10%和质量值Q≤10的碱基数占整条Read50%以上的Reads),获得可后续分析的高质量Clean Data。利用HISAT2系统[18]将Clean Data与棉蚜基因组数据(NCBI: ASM2018417v2)进行比对,获得Mapped data。随后,通过StringTie软件 [19]对Mapped data进行转录组组装。最后,通过BLAST[20]在Non-redundant protein sequences(NR) [21],Swiss-Prot [22],Gene ontology(GO)[23],Database of clusters of orthologous genes(COG)[24],KOG[25],Protein families database(Pfam)[26],Kyoto encyclopedia of genes and genomes(KEGG)[27]和Evolutionary genealogy of genes: Non-supervised orthologous groups databases(eggNOG)[28]数据库进行序列比对,获得功能注释。
1.2.3 基因差异表达
标准化样品中Mapped Reads数目和转录本长度,并以FPKM(Fragments per kilobase of transcript per million fragments mapped)[29]作为基因表达水平指标。通过DESeq2[30]对样品组间进行差异表达分析,以log2 Fold change ≥ 1且FDR(False Discovery Rate)lt; 0.05作为筛选差异表达基因的标准,并对差异表达基因进行GO功能分析。
2 结果与分析
2.1 测序数据评估
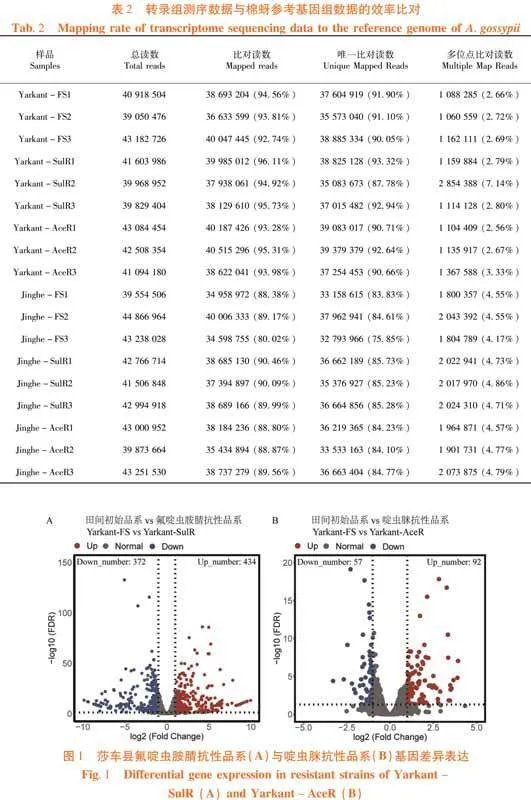
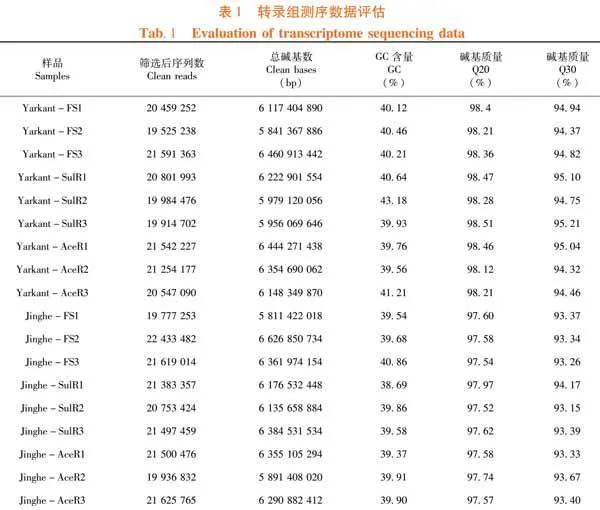
研究表明,测序数据经过去除含有接头序列和低质量序列,莎车县田间初始品系、氟啶虫胺腈抗性品系和啶虫脒抗性品系9个样品共获得55.53 Gb Clean Data,各样品Clean Data均达到5.84 Gb,Q30碱基百分比均在94.32%以上,共计185 620 518个Clean reads;精河县田间初始品系、氟啶虫胺腈抗性品系和啶虫脒抗性品系9个样品共获得56.03 Gb Clean Data,各样品Clean Data均达到5.81 Gb,Q30碱基百分比在93.15%以上,共计190 527 062个Clean reads。表1
2.2 测序数据与参考基因组比对效率
研究表明,与棉蚜参考基因组序列(NCBI: ASM2018417v2)比对,莎车县3个品系9个样品的Clean Reads对比效率在92.74% ~ 96.11%,唯一比对读数占总读数比例在87.78% ~ 92.94%,多位点比对度数占总读数比例在2.56% ~ 7.14%;精河县3个品系9个样品的Clean Reads对比效率在80.02% ~ 90.46%,唯一比对读数占总读数比例在75.85% ~ 85.73%,多位点比对度数占总读数比例在4.17% ~ 4.86%。表2
2.3 差异表达基因
研究表明,莎车县氟啶虫胺腈品系相对其田间初始品系共有806条基因差异表达,其中434条基因显著性上调表达,372条基因显著性下调表达(图1A);啶虫脒抗性品系相对其田间初始品系共有149条基因差异表达,其中92条基因显著性上调表达,57条基因显著性下调表达(图1B)。图1
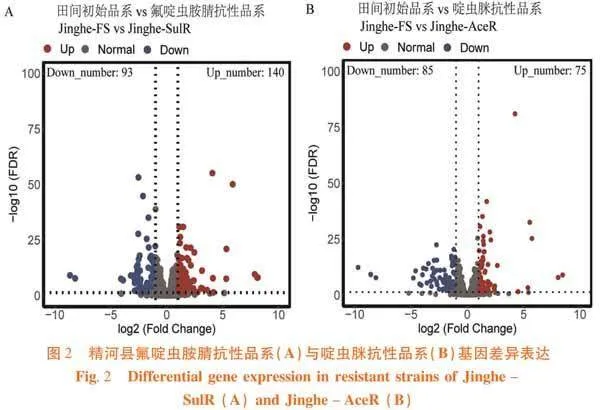
精河县氟啶虫胺腈品系相对其田间初始品系共有233条基因差异表达,其中140条基因显著性上调表达,93条基因显著性下调表达(图2A);啶虫脒抗性品系相对其田间初始品系共有160条基因差异表达,其中有75条基因显著性上调表达,85条基因显著性下调表达(图2B)。图2
2.4 差异表达基因GO功能分类
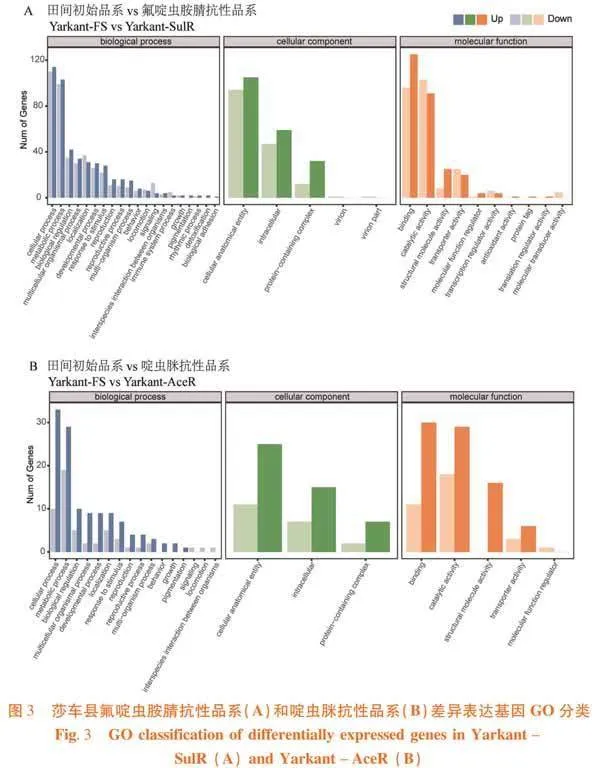
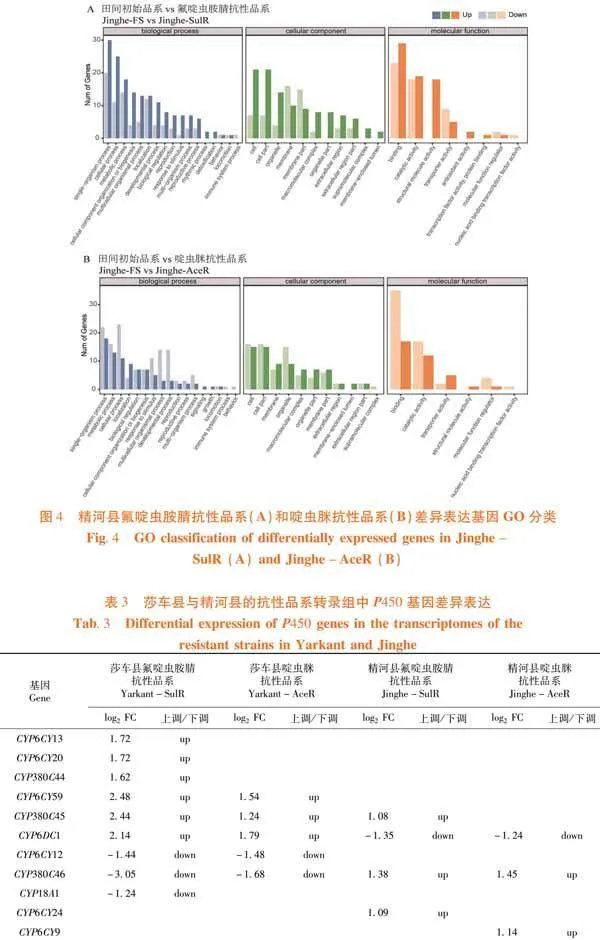
研究表明,莎车县氟啶虫胺腈抗性品系和啶虫脒抗性品系差异表达基因经GO功能分类,参与生物过程(biological process)的差异表达基因数目最多,其次为分子功能(molecular function),最少为细胞组件(cellular component)。与莎车县田间初始品系相比,莎车县氟啶虫胺腈抗性品系在生物过程中差异基因888个,主要分布于细胞过程(cellular process)(上调114个,下调110个)和代谢过程(metabolic process)(上调103个,下调99个);在分子功能中,差异基因516个,主要分布于细胞结合(binding)(上调125个,下调96个)和催化活性(catalytic activity)(上调91个,下调103个);在细胞组件中,差异基因351个,主要分布于细胞解剖实体(cellular anatomical entity)(上调105个,下调94个)和细胞内(intracellular)(上调59个,下调47个)(图3A)。莎车县啶虫脒抗性品系在生物过程中差异基因175个,主要分布于代谢过程(上调29个,下调19个)和细胞过程(上调33个,下调10个);在分子功能中,差异基因114个,主要分布于细胞结合(上调30个,下调11个)和催化活性(上调29个,下调18个);在细胞组件中,差异基因67个,主要分布于细胞解剖实体(上调25个,下调11个)和细胞内(上调15个,下调7个)(图3B)。图3
精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系差异表达基因经GO功能分类,差异表达基因数目由高到低依次为生物过程、细胞组件和分子功能。与精河县田间初始品系相比,精河县氟啶虫胺腈抗性品系在生物过程中差异表达基因166个,主要分布于单生物过程(single-organism process)(上调30个,下调20)和细胞过程(cellular process)(上调25个,下调11);细胞组件中差异表达基因166个,主要分布于膜(membrane)(上调10个,下调16个)和细胞(Cell)(上调21个,下调7个);分子功能差异表达基因128个,主要分布于细胞结合(上调29个,下调23个)和催化活性(上调19个,下调18个)(图4A)。精河县啶虫脒抗性品系生物过程中差异表达基因217个,主要分布于单生物过程(上调18个,下调22)和细胞过程(上调11个,下调23);细胞组件中差异表达基因149个,主要分布于细胞部分(cell part)(上调15个,下调16个)和细胞器(organelle)(上调9个,下调15个);分子功能差异表达基因95个,主要分布于细胞结合(上调17个,下调35个)和催化活性(上调12个,下调17个)(图4B)。图4
2.5 莎车县与精河县各抗性品系转录组中P450基因的表达差异
研究表明,与莎车县田间初始品系相比,莎车县氟啶虫胺腈抗性品系显著上调和下调表达的P450基因数目均多于啶虫脒抗性品系。氟啶虫胺腈抗性品系中,6个P450基因显著上调表达,即:CYP6CY59、CYP6CY20、CYP6DC1、CYP6CY13、CYP380C44和CYP380C45;3个P450基因显著下调表达,即:CYP18A1、CYP6CY12和CYP380C46。啶虫脒抗性品系中,3个P450基因显著上调表达,即:CYP6CY59、CYP6DC1和CYP380C45;2个P450基因显著下调表达,即:CYP6CY12和CYP380C46。与精河县田间初始品系相比,精河县氟啶虫胺腈抗性品系显著上调和下调表达的P450基因数目均多于啶虫脒抗性品系。氟啶虫胺腈抗性品系中,3个P450基因显著上调表达,即:CYP6CY24、CYP380C46和CYP380C45;1个P450基因显著下调表达,即:CYP6DC1。啶虫脒抗性品系中,CYP6CY9和CYP380C46显著上调表达,CYP6DC1显著下调表达。表3
3 讨 论
3.1
不同地域的环境、害虫管理和杀虫剂使用习惯的不同,包括杀虫剂种类、施药量和施药频率,使得不同地域棉蚜田间种群对新烟碱类杀虫剂抗性存在差异[31],不同地域棉蚜种群具有不同的抗性抗性遗传背景。啶虫脒与氟啶虫胺腈都是烟碱型乙酰胆碱受体激动剂,然而氟啶虫胺腈作用机制有别于啶虫脒等新烟碱类杀虫剂[32]。
相同抗性遗传背景下,氟啶虫胺腈抗性品系差异表达基因数量要多于啶虫脒抗性品系。莎车县氟啶虫胺腈品系和啶虫脒抗性品系分别有806个(上调434个,下调372个)和149个(上调92个,下调57个)基因差异表达,精河县氟啶虫胺腈品系和啶虫脒抗性品系分别有233个(上调140个,下调93个)和160个(上调75个,下调85个)基因差异表达。产生关于杀虫剂抗性和关键生活史性状的能量权衡[33-35]。由于莎车县和精河县氟啶虫胺腈抗性品系差异表达基因均多于啶虫脒抗性品系,氟啶虫胺腈品系可能伴随着更高的能量消耗,这种能量的高消耗导致了氟啶虫胺腈抗性品系适合度低于啶虫脒抗性品系[36]。
3.2
经GO功能分析,莎车县、精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系差异表达基因在生物过程、分子功能和细胞组件中分布不同。莎车县氟啶虫胺腈抗性品系和啶虫脒抗性品系差异表达基因分布由高到低依次为生物过程、分子功能和细胞组件,而精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系差异表达基因分布由高到低依次为生物过程、细胞组件和分子功能。除此之外,在生物过程、分子功能和细胞组件中的具体分布亦有所不同。莎车县氟啶虫胺腈抗性品系和啶虫脒抗性品系差异表达基因在生物过程中主要分布于细胞过程和代谢过程,在分子功能中主要分布于细胞结合和催化活性,在细胞组件中主要分布于细胞解剖实体和细胞内。精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系差异表达基因在生物过程中差异表达基因主要分布于单生物过程和细胞过程,分子功能差异表达基因主要分布于细胞结合和催化活性。在细胞组件中,精河县氟啶虫胺腈抗性品系差异表达基因主要分布于膜和细胞,而啶虫脒抗性品系差异表达基因主要分布于细胞部分和细胞器。
3.3
细胞色素P450多功能氧化酶是害虫重要的解毒酶。以往大量研究已经证实棉蚜[5,37,38]、桃蚜[39]、烟粉虱[40]和褐飞虱[41]等害虫对新烟碱类杀虫剂抗性发展与部分P450基因过量表达有关。基于转录组数据,研究对莎车县和精河县棉蚜种群的P450相关基因进行了筛选。在4个抗性品系共涉及差异表达P450相关基因11个,莎车县氟啶虫胺腈抗性品系和啶虫脒抗性品系差异表达P450相关基因分别为9个(6个上调,3个下调)和5个(3个上调,2个下调),精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系差异表达P450相关基因分别为4个(3个上调,1个下调)和3个(2个上调,1个下调)。莎车县和精河县氟啶虫胺腈抗性品系显著上调表达P450相关基因数量多于2个啶虫脒抗性品系,而且莎车县氟啶虫胺腈抗性品系和啶虫脒抗性品显著上调表达P450相关基因数量均分别多于精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系。此外,CYP380C45均在莎车县氟啶虫胺腈抗性品系、莎车县啶虫脒抗性品系和精河县氟啶虫胺腈抗性品系中显著上调表达。CYP380C46在莎车县氟啶虫胺腈抗性品系和啶虫脒抗性品系中均显著下调表达,却在精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系均显著上调表达。相反,CYP6DC1在莎车县氟啶虫胺腈抗性品系和啶虫脒抗性品系中均显著上调表达,却在精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系均显著下调表达。莎车县和精河县棉蚜对氟啶虫胺腈抗性发展要快于啶虫脒,莎车县2个抗性品系抗性发展快于精河县2个抗性品系[36],可能与上述P450相关基因在地域和品系之间表达差异有关。
4 结 论
有多个P450基因参与棉蚜对氟啶虫胺腈和啶虫脒的抗性。差异表达的P450基因在不同抗性品系间存在差异,而且在不同抗性遗传背景棉蚜种群之间也存在差异。相同抗性遗传背景下,氟啶虫胺腈抗性品系差异表达P450基因数量多于啶虫脒抗性品系;不同抗性遗传背景下,莎车县氟啶虫胺腈抗性品系和啶虫脒抗性品系差异表达P450基因数量均分别多于精河县氟啶虫胺腈抗性品系和啶虫脒抗性品系。
参考文献(References)
[1]热依汗古丽·阿布都热合曼, 艾合买提·吾斯曼, 魏新政, 等. 2019年新疆棉花主要病虫害发生概况[J]. 中国棉花, 2019, 46(11): 7-9.
Reyihanguli Abudureheman, Aihemaiti Wusiman, WEI Xinzheng, et al. Overview of main cotton diseases and insect pests in Xinjiang in 2019[J]. China Cotton, 2019, 46(11): 7-9.
[2] 热依汗古丽·阿布都热合曼, 伊力亚尔·达吾提江, 艾合买提江·努力买买提, 等. 2020年新疆棉花主要病虫害发生概况[J]. 中国棉花, 2021, 48(2): 10-12, 16.
Reyihanguli Abudureheman, Yiliyaer Dawutijiang, Aihemaitijiang Nulimaimaiti, et al. Overview of main cotton diseases and insect pests in Xinjiang in 2020[J]. China Cotton, 2021, 48(2): 10-12, 16.
[3] 帕提玛·乌木尔汗, 郭佩佩, 马少军, 等. 新疆地区棉蚜田间种群对10种杀虫剂的抗性[J]. 植物保护, 2019, 45(6): 273-278.
Patima Wumuerhan, GUO Peipei, MA Shaojun, et al. Resistance of different field populations of Aphis gossypii to ten insecticides in Xinjiang[J]. Plant Protection, 2019, 45(6): 273-278.
[4] 马康生, 王静慧, 解晓平, 等. 棉蚜对新烟碱类杀虫剂的抗性现状及其治理策略[J]. 植物保护学报, 2021, 48(5): 947-957.
MA Kangsheng, WANG Jinghui, XIE Xiaoping, et al. Status and management strategies of neonicotinoid insecticide resistance in Aphis gossypii Glover[J]. Journal of Plant Protection, 2021, 48(5): 947-957.
[5] Zhang H H, Yang H L, Dong W Y, et al. Mutations in the nAChR β1 subunit and overexpression of P450 genes are associated with high resistance to thiamethoxam in melon aphid, Aphis gossypii Glover[J]. Comparative Biochemistry and Physiology Part B, Biochemistry amp; Molecular Biology, 2022, 258: 110682.
[6] Chen X W, Xia J, Shang Q L, et al. UDP-glucosyltransferases potentially contribute to imidacloprid resistance in Aphis gossypii glover based on transcriptomic and proteomic analyses[J]. Pesticide Biochemistry and Physiology, 2019, 159: 98-106.
[7] Pan Y O, Zeng X C, Wen S Y, et al. Multiple ATP-binding cassette transporters genes are involved in thiamethoxam resistance in Aphis gossypii glover[J]. Pesticide Biochemistry and Physiology, 2020, 167: 104558.
[8] Bass C, Denholm I, Williamson M S, et al. The global status of insect resistance to neonicotinoid insecticides[J]. Pesticide Biochemistry and Physiology, 2015, 121: 78-87.
[9] Hirata K, Jouraku A, Kuwazaki S, et al. The R81T mutation in the nicotinic acetylcholine receptor of Aphis gossypii is associated with neonicotinoid insecticide resistance with differential effects for cyano- and nitro-substituted neonicotinoids[J]. Pesticide Biochemistry and Physiology, 2017, 143: 57-65.
[10] Kim J I, Kwon M, Kim G H, et al. Two mutations in nAChR beta subunit is associated with imidacloprid resistance in the Aphis gossypii[J]. Journal of Asia-Pacific Entomology, 2015, 18(2): 291-296.
[11] Chen X W, Li F, Chen A Q, et al. Both point mutations and low expression levels of the nicotinic acetylcholine receptor β1 subunit are associated with imidacloprid resistance in an Aphis gossypii (Glover) population from a Bt cotton field in China[J]. Pesticide Biochemistry and Physiology, 2017, 141: 1-8.
[12] Shi X G, Zhu Y K, Xia X M, et al. The mutation in nicotinic acetylcholine receptor β1 subunit may confer resistance to imidacloprid in Aphis gossypii (Glover)[J]. Journal of Food, Agriculture and Environment, 2012, 10(2): 1227-1230.
[13] Koo H N, An J J, Park S E, et al. Regional susceptibilities to 12 insecticides of melon and cotton aphid, Aphis gossypii (Hemiptera: Aphididae) and a point mutation associated with imidacloprid resistance[J]. Crop Protection, 2014, 55: 91-97.
[14] Wei X, Pan Y O, Xin X C, et al. Cross-resistance pattern and basis of resistance in a thiamethoxam-resistant strain of Aphis gossypii Glover[J]. Pesticide Biochemistry and Physiology, 2017, 138: 91-96.
[15] IRAC. The IRAC mode of action classification online [J/OL] 2023, [2023-4-12]. https://irac-online.org/mode-of-action/classification-online.
[16] Babcock J M, Gerwick C B, Huang J X, et al. Biological characterization of sulfoxaflor, a novel insecticide[J]. Pest Management Science, 2011, 67(3): 328-334.
[17] Cutler P, Slater R, Edmunds A J F, et al. Investigating the mode of action of sulfoxaflor: a fourth-generation neonicotinoid[J]. Pest Management Science, 2013, 69(5): 607-619.
[18] Kim D, Langmead B, Salzberg S L. HISAT: a fast spliced aligner with low memory requirements[J]. Nature Methods, 2015, 12(4): 357-360.
[19] Pertea M, Pertea G M, Antonescu C M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads[J]. Nature Biotechnology, 2015, 33(3): 290-295.
[20] Altschul S F, Madden T L, Schffer A A, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs[J]. Nucleic Acids Research, 1997, 25(17): 3389-3402.
[21] 邓泱泱, 荔建琦, 吴松锋, 等. nr数据库分析及其本地化[J]. 计算机工程, 2006, 32(5): 71-73, 76.
DENG Yangyang, LI Jianqi, WU Songfeng, et al. Integrated nr database in protein annotation system and its localization[J]. Computer Engineering, 2006, 32(5): 71-73, 76.
[22] The UniProt Consortium. UniProt: the universal protein knowledgebase[J]. Nucleic Acids Research, 2017, 45(D1): D158-D169.
[23] Ashburner M, Ball C A, Blake J A, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium[J]. Nature Genetics, 2000, 25(1): 25-29.
[24] Tatusov R L, Galperin M Y, Natale D A, et al. The COG database: a tool for genome-scale analysis of protein functions and evolution[J]. Nucleic Acids Research, 2000, 28(1): 33-36.
[25] Koonin E V, Fedorova N D, Jackson J D, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes[J]. Genome Biology, 2004, 5(2): R7.
[26] Punta M, Coggill P C, Eberhardt R Y, et al. The Pfam protein families database[J]. Nucleic Acids Research, 2012, 40(Database issue): D290-D301.
[27] Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome[J]. Nucleic Acids Research, 2004, 32(Database issue): D277-D280.
[28] Huerta-Cepas J, Szklarczyk D, Heller D, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses[J]. Nucleic Acids Research, 2019, 47(D1): 309-314.
[29] Florea L, Song L, Salzberg S L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues[J]. F1000Research, 2013,2: 188.
[30] Love M I, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biology, 2014, 15(12): 550.
[31] 赵鹏程, 李焱, 闫文静, 等. 新疆棉蚜不同地理种群对杀虫剂的敏感性[J]. 石河子大学学报(自然科学版), 2018, 36(2): 159-163.
ZHAO Pengcheng, LI Yan, YAN Wenjing, et al. Sensitivity of different geographical populations of Aphis gossypii(Glover) in Xinjiang to different insecticides[J]. Journal of Shihezi University (Natural Science), 2018, 36(2): 159-163.
[32] Oliveira E E, Schleicher S, Büschges A, et al. Desensitization of nicotinic acetylcholine receptors in central nervous system neurons of the stick insect (Carausius morosus) by imidacloprid and sulfoximine insecticides[J]. Insect Biochemistry and Molecular Biology, 2011, 41(11): 872-880.
[33] ffrench-Constant R H, Bass C. Does resistance really carry a fitness cost?[J]. Current Opinion in Insect Science, 2017, 21: 39-46.
[34] Roush R T, McKenzie J A. Ecological genetics of insecticide and acaricide resistance[J]. Annual Review of Entomology, 1987, 32: 361-380.
[35] Rivero A, Magaud A, Nicot A, et al. Energetic cost of insecticide resistance in Culex pipiens mosquitoes[J]. Journal of Medical Entomology, 2011, 48(3): 694-700.
[36] Wang W, Zhang R F, Liu H Y, et al. Resistance development, cross-resistance, and fitness costs associated with Aphis gossypii resistance towards sulfoxaflor and acetamiprid in different geographical regions[J]. Journal of Integrative Agriculture, 2024, 23(7): 2332-2345.
[37] Zhao L K, Wang C P, Gao X K, et al. Characterization of P450 monooxygenase gene family in the cotton aphid, Aphis gossypii Glover[J]. Journal of Asia-Pacific Entomology, 2022, 25(2): 101861.
[38] Wang L, Cui L, Wang Q Q, et al. Sulfoxaflor resistance in Aphis gossypii: resistance mechanism, feeding behavior and life history changes[J]. Journal of Pest Science, 2022, 95(2): 811-825.
[39] Pym A, Umina P A, Reidy-Crofts J, et al. Overexpression of UDP-glucuronosyltransferase and cytochrome P450 enzymes confers resistance to sulfoxaflor in field populations of the aphid, Myzus persicae[J]. Insect Biochemistry and Molecular Biology, 2022, 143: 103743.
[40] He C, Liang J J, Liu S N, et al. Molecular characterization of an NADPH cytochrome P450 reductase from Bemisia tabaci Q: potential involvement in susceptibility to imidacloprid[J]. Pesticide Biochemistry and Physiology, 2020, 162: 29-35.
[41] Cheng Y B, Li Y M, Li W R, et al. Inhibition of hepatocyte nuclear factor 4 confers imidacloprid resistance in Nilaparvata lugens via the activation of cytochrome P450 and UDP-glycosyltransferase genes[J]. Chemosphere, 2021, 263: 128269.
Transcriptome analysis of Aphis gossypii sulfoxaflor and acetamiprid-resistant strains with different genetic backgrounds of resistance
WANG Wei1, 2, ZHANG Renfu1, LIU Haiyang1, DING Ruifeng1, LIANG Gemei2, YAO Ju1
(1. Key Laboratory of Integrated Pest Management on Crop in Northwestern Oasis, Ministry of Agriculture and Rural Affairs/ Xinjiang Key Laboratory of Agricultural Biosafety /Institute of Plant Protection,Xinjiang Academy of Agricultural Sciences,Urumqi 830091,China;2. State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China)
Abstract:【Objective】 To explore the differences in resistance mechanisms to sulfoxaflor and acetamiprid in cotton aphid with different genetic backgrounds of resistance.
【Methods】 Transcriptome sequencing by Illumina high-throughput sequencing technology was performed on initial field strain, acetamiprid-resistant strain and sulfoxaflor-resistant strain of cotton aphids with different genetic backgrounds (Yarkant and Jinghe), respectively.Meanwhile, differentially expressed genes in resistant strains of cotton aphid with different genetic backgrounds were analyzed by bioinformatics methods.
【Results】 By comparing the transcriptome data of sulfoxaflor- and acetamiprid-resistant strains from Yarkant and Jinghe, it was found that the sulfoxaflor- and acetamiprid-resistant strains from Yarkant had differential expression of 806 and 149 genes, respectively, and the sulfoxaflor- and acetamiprid-resistant strains from Jinghe had differential expression of 233 and 160 genes.In the sulfoxaflor- and acetamiprid-resistant strains of Yarkant, CYP6CY59, CYP6DC1, and CYP380C45 were up-regulated, but CYP6CY12 and CYP380C46 were down-regulated.In the sulfoxaflor- and acetamiprid-resistant strains of Jinghe, CYP380C46 was up-regulated whereas CYP6DC1 was down-regulated.In addition, CYP380C45 was up-regulated in Yarkant sulfoxaflor- and acetamiprid-resistant strains, and the Jinghe acetamiprid-resistant strain.CYP6DC1 was up-regulated in both Yarkant resistant strains, but down-regulated in both Jinghe resistant strains.CYP380C46 was up-regulated in both resistant strains in Jinghe but down-regulated in both resistant strains in Yarkant.
【Conclusion】" Several P450 genes were involved in resistance to sulfoxaflor and acetamiprid in cotton aphids.Differences in differentially expressed P450 genes were found between sulfoxaflor- and acetamiprid-resistant strains of cotton aphids with the same genetic background, and found between sulfoxaflor-resistant strains of cotton aphids of different genetic backgrounds, as well as between acetamiprid-resistant strains.
Key words: Aphis gossypii; resistance; differentially expressed genes
Fund projects:National Key Ramp;D Program of China (2022YFD1400300); Project for Stable Support to Agricultural Sci - Tech Renovation (xjnkywdzc-2023004-1)
Correspondence author:YAO Ju (1969-), male, from Shandong, researcher, bachelors degree, research direction: integrated pest management in cotton, (E-mail) yaoju500@sohu.com
LIANG Gemei (1970-), female," from Beijing, researcher, doctors degree, research direction: integrated pest management in cotton, (E-mail) gmliang@ippcaas.cn
基金项目:国家重点研发计划(2022YFD1400300);农业科技创新稳定支持项目(xjnkywdzc-2023004-1)
作者简介:王伟(1982-),男,天津人,研究员,博士,研究方向为棉花有害生物防治,(E-mail)wlzforever2004@sina.com
通讯作者:姚举(1969-),男,山东人,研究员,硕士,研究方向为棉花有害生物防治,(E-mail)yaoju500@sohu.com
梁革梅(1970-),女,北京人,研究员,博士,研究方向为棉花有害生物防治,(E-mail)gmliang@ippcaas.cn