doi:10.6048/j.issn.1001-4330.2024.05.028
摘" 要:【目的】研究海南猪繁殖与呼吸综合征病毒PRRSV的流行病学特征和遗传变异规律。
【方法】采集海南省海口市及周边地区样品,分离出PRRSV HNHK3-2021 毒株,并测定NSP2基因以及 ORF5 和ORF7基因序列,采用Lasergene 7.1进行序列比对分析,用MEGA-X进行遗传演化分析。
【结果】分离出的PRRSV HNHK3-2021株为美洲型毒株,PRRSV HNHK3-2021株和EF112447 HEB1、EU109503 GD、EU624117 XH-GD的ORF5和ORF7的核苷酸相似性较高,分别为99.2%~99.3%和99.5%~100%,推导氨基酸相似性为99%~99.5%和100%;PRRSV HNHK3-2021株和EU864232 SHB、AY262352_HB-2(sh)2002、AY656990 Abst-1、AY032626 CH-1a、EU807840 CH-1R的ORF5和ORF7的核苷酸相似性为87.1%~96.4%和91.4%~97.3%,推导氨基酸的相似性为86.6%~93%和91.9%~97.6%;PRRSV HNHK3-2021株和JN654459 NADC30、KP860909 FJZ03、MH068878 SD17-38、AY881994 FJ-1的ORF5和ORF7的核苷酸相似性为85.4%~87.1%和89%~91.4%。氨基酸的相似性为86.1%~87.1%和89.5%~91.9%。
【结论】RRSV HNHK3-2021 毒株和EU624117 XH-GD具有最近的遗传距离。
关键词:
猪繁殖与呼吸综合征;遗传进化;ORF5;ORF7
中图分类号:S85""" 文献标志码:A""" 文章编号:1001-4330(2024)05-1292-09
收稿日期(Received):
2023-10-12
基金项目:
海南省省属科研院所技术创新专项(jscx202001);国家自然科学基金项目(32060796);家畜疫病病原生物学国家重点实验室开放基金(SKLVEB2020KFKT021);海南省农业科学院院本级科研项目(HAAS2022PT0205);国家重点研发计划(2023YFC3404302)
作者简介:
范悦轩(1997-),男,新疆奇台人,硕士研究生,研究方向为家畜传染病学,(E-mial)1245219231@qq.com
通讯作者:
曹宗喜(1982-) ,男,河南新乡人,研究员,博士,硕士生/博士生导师,研究方向为家畜传染病学,(E-mial)caozongxi@163.com
刘光亮(1975-),男,四川资中人,研究员,博士,硕士生/博士生导师,研究方向为动物疫苗研制及分子免疫学,(E-mial)liuguangliang01@caas.cn
0" 引 言
【研究意义】 猪繁殖与呼吸综合征(Porcine reproductive and respiratory syndrome,PRRS)是由猪繁殖与呼吸综合征病毒(PRRS virus,PRRSV)所引起的一种高度接触性传染病,引起母猪流产和仔猪呼吸道疾病。通过对海南省海口市及周边地区样品的分离鉴定后,分析典型毒株PRRSV HNHK3-2021的基因,了解PRRSV在海南的流行情况,进而对海南PRRS防控提供支持。【前人研究进展】猪繁殖与呼吸综合征病毒PRRSV危害猪养殖业[1]。PRRSV为单股正链RNA病毒,长度大约为15 kb,至少包含1个开放阅读框(ORF1a、ORF1b、ORF2a、ORF2b、ORF3、ORF4、ORF5、ORF5a、ORF6、ORF7),NSP2作为PRRSV最大的复制酶蛋白[2],基于各种亚型的PRRSV在该区域差异极为明显[3]。NSP2能够顺式或反式切割 Nsp2/3结合处[4],在PRRSV的复制中 至关重要的作用[5]。ORF5编码的囊膜蛋白是 GP5,GP5含有与病毒中和、病毒保护相关的表位[6]。ORF5基因是 PRRSV 基因组中变化最大的区域之一,各个亚型的PRRSV在此处同样差异明显,基于ORF5的PRRSV的分型是公认的PRRSV分型[7]。ORF7编码的N蛋白是病毒的核衣壳蛋白[8],该蛋白在血清学反应中较为保守[9]。【本研究切入点】有关海南猪繁殖与呼吸综合征和PRRSV的研究数据较少,
需对毒株PRRSV HNHK3-2021的 ORF5 和ORF7基因进行测序分析。【拟解决的关键问题】
对海南省海口市及周边地区采样,进行病毒分离,并对分离获得的毒株PRRSV HNHK3-2021进行NSP2的基因以及 ORF5 和ORF7基因序列测定分析,了解海南PRRS分子流行情况。
1" 材料与方法
1.1" 材 料
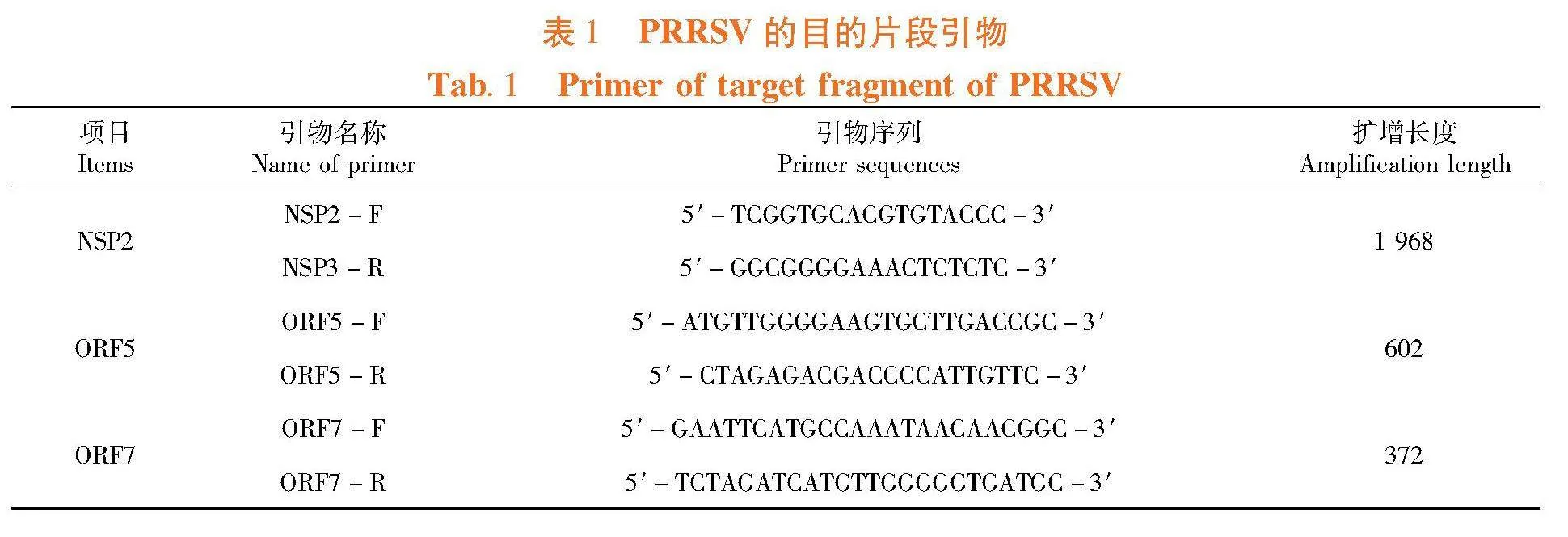
Marc-145细胞由海南省热带动物繁育与疫病研究重点实验室保存;Trizol 试剂、胎牛血清、DMEM培养基、购自赛默飞世尔科技(中国)有限公司Ex Taq DNA 聚合酶、pMD18 -T购自宝生物工程(大连)有限公司,Plasmid Mini Kit、Gel DNA Exteration Mini Kit、反转录酶购自南京诺唯赞生物科技有限公司。NSP2、ORF5、ORF7的扩增引物参照贺冬梅等[10]。表1
1.2" 方 法
1.2.1" 样品中基因提取
用研钵将组织样研磨碎,使用Trizol法提取RNA,使用反转录试剂盒对样品反转录,反转录体系为5× H:SCRIPT Ⅱ qFT Super Mix Ⅱ 4 μL, RNA 100 ng ,dd水补齐至20 μL。反应条件为50℃ 15 min;85℃ 5 s。反转录产物进行cDNA的PCR,将PCR产物进行琼脂糖凝胶电泳,条件为恒压90 V,电泳结束后使用紫外凝胶成像系统观察试验结果,筛选出阳性样品。
1.2.2" 病原分离
将筛选出的阳性样品接种到Marc-145细胞上,放入温箱每天观察细胞病变情况,细胞病变达60%~70%,将细胞反复冻融3次,4℃" 6 000 r/min离心10 min,取上清。如此反复,盲传3~5代。
将已经盲传3~5代,有明显病变的Marc-145放入-80℃冰箱,直至完全冻结后在室温完全溶解,如此反复3次后,提取RNA,反转录,进行PCR检测。
1.2.3" 扩增目的基因
病毒RNA采用Trizol法提取,反转录体系为5× H:SCRIPT Ⅱ qFT Srper Mix Ⅱ 4 μL, RNA 100 ng ,dd水补齐至20 μL。反应条件为50℃ 15 min;85℃ 5 s。PRC反应体系:。Ex Taq DNA 聚合酶预混剂25 μL,底物2 μL,水23 μL。反应条件:94℃ 3 min;94℃ 10 s,55℃ 30 s,72℃ 2 min,共30个循环[11]。
1.2.4" 目的基因的连接转化
将PCR产物进行琼脂糖凝胶电泳,对目的条带进行胶回收,回收产物和pMD18 -T连接,连接产物转化于DH5α感受态细胞。37℃温箱培养16 h后挑取阳性菌落送测序。
1.2.5" 测序
从NCBI选取AY032626 CH-1a等14株PRRSV序列运用Lasergene 7.1进行序列比对和相似性分析,用MEGA-X进行遗传演化分析对测序结果进行序列比对。
2" 结果与分析
2.1" 病原分离结果
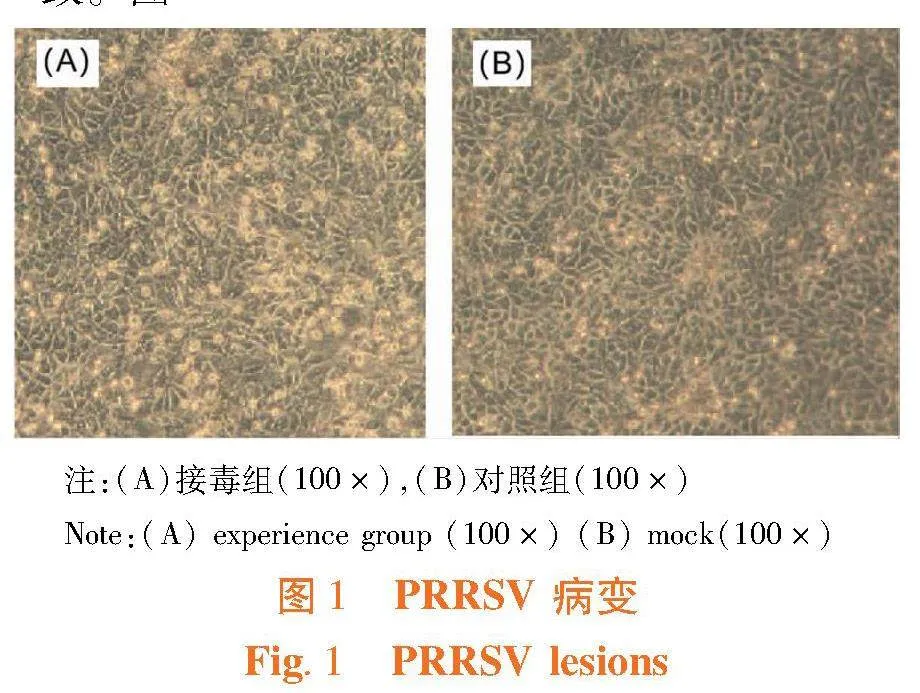
研究表明,检测为PRRSV阳性的样品接种到Mrac-145细胞上,培养7 d,盲传3代后,出现细胞圆缩、空泡化、折光性增强、聚集、脱落的细胞病变。2021年度成功分离出6株PRRSV毒株。盲传3代后,将出现病变的细胞通过反复冻融的方法收集病毒,提取RNA后反转录为得到cDNA,使用PCR方法对得到的cDNA进行复检验证结果一致。图1
2.2" NSP2分析
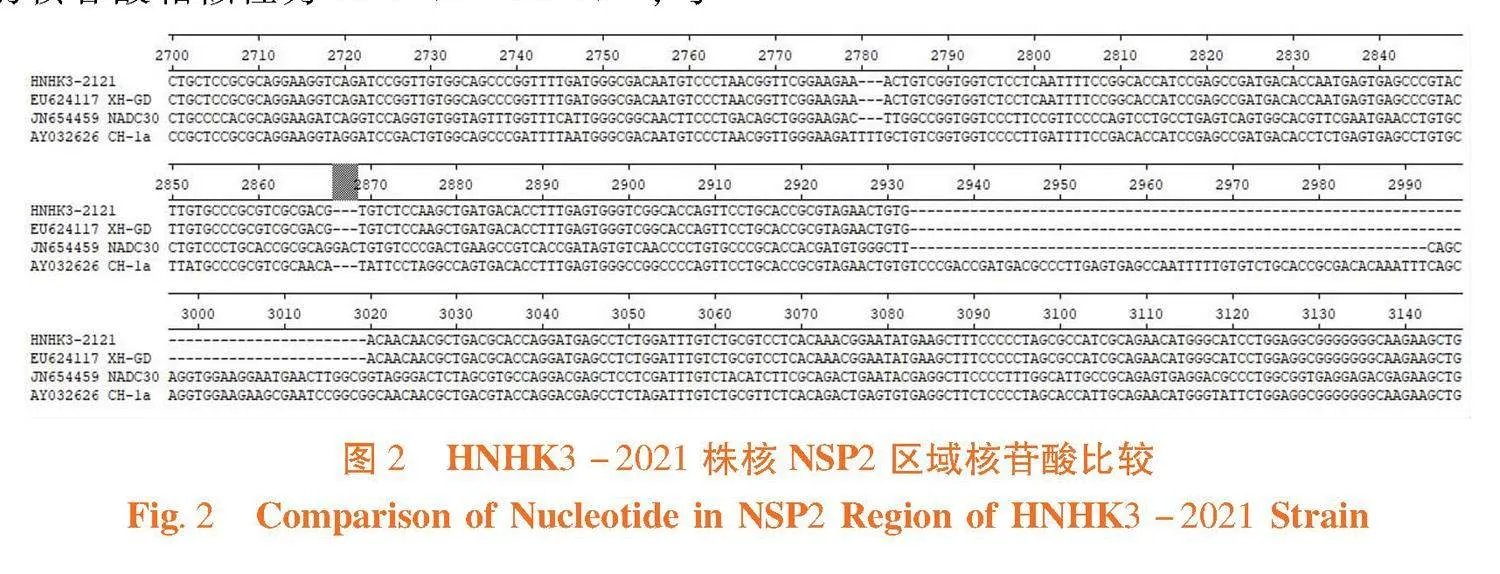
研究表明,对比PRRSV HNHK3-2021株核酸序NSP2与其他毒株,其相较于经典株AY032626 CH-1a在2 780~2 712和2 887~2 992发生了不连续缺失,与高致病毒株EU624117 XH-GD变异株的缺失位置一致,PRRSV HNHK3-2021属于美洲型高致病型毒株。图1
2.3" ORF5基因序列分析
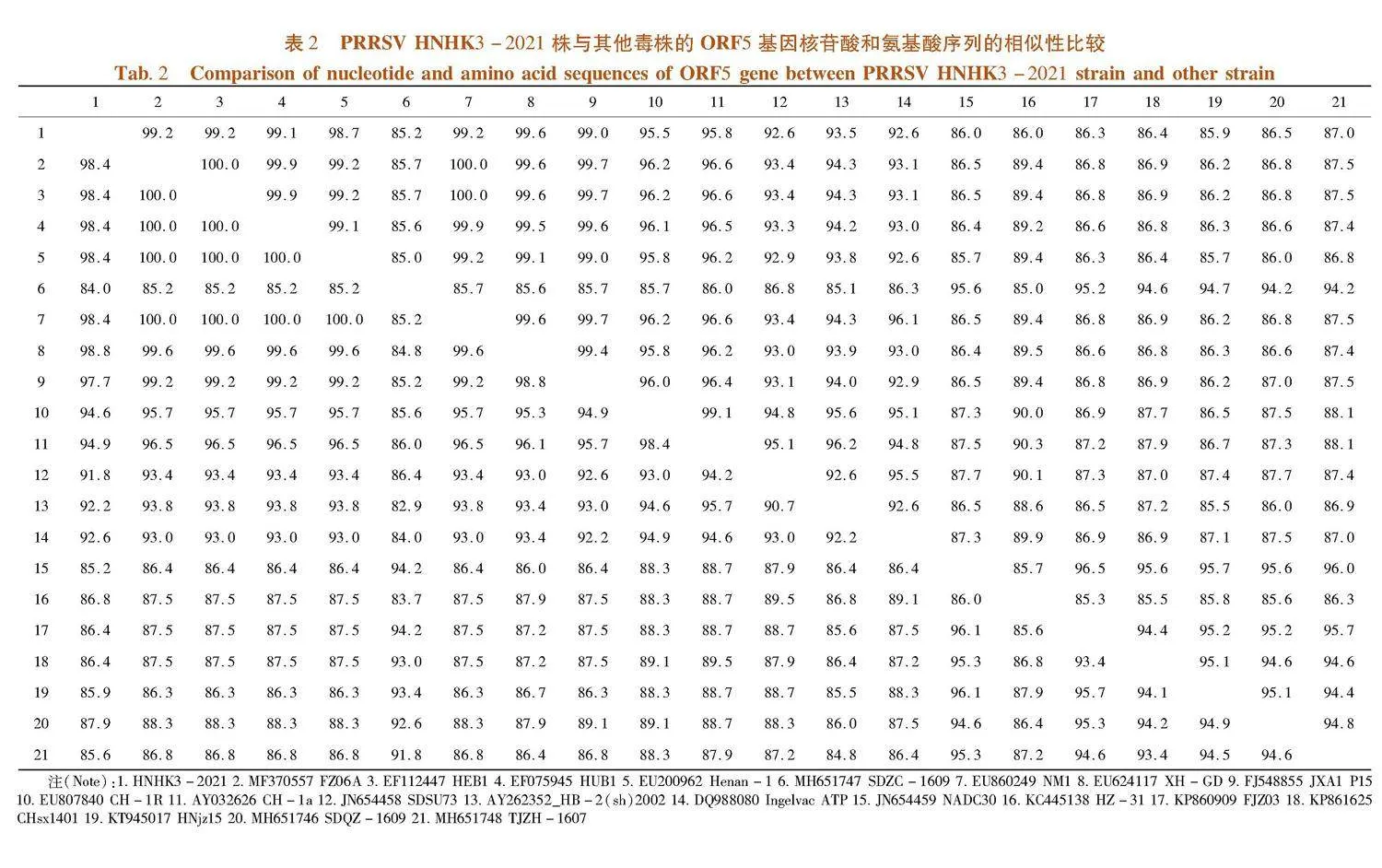
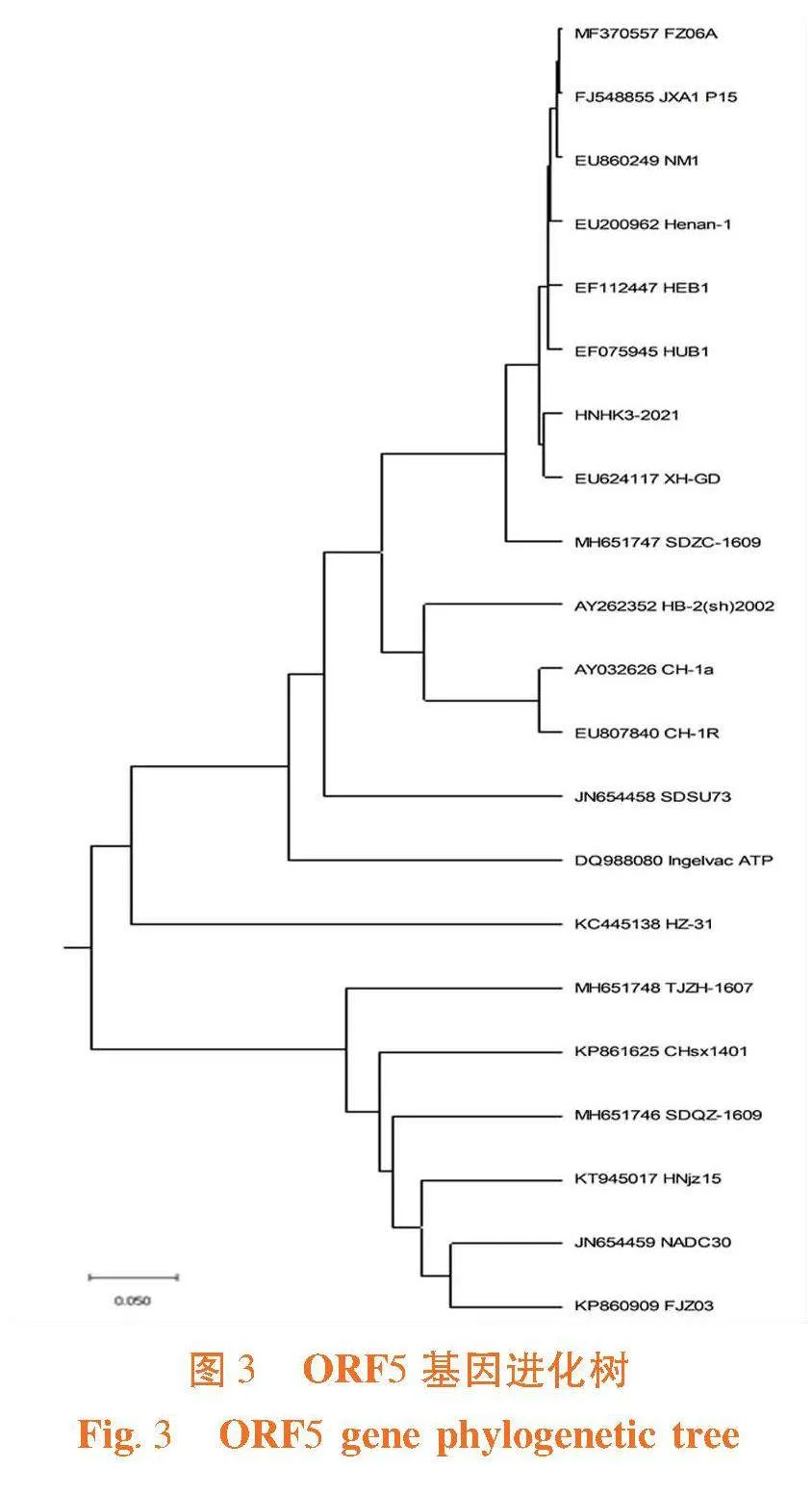
研究表明,ORF5基因片段的序列为603 bp,PRRSV HNHK3-2021株的ORF5和EF112447 HEB1、MF370557 FZ06A、EF075945 HUB1、FJ548855 JXA1 P15、EU200962、 Henan-1、EU624117 XH-GDDE等高致性PRRSV的等位基因的核苷酸相似性为98.72%~99.3% ;与KC445138 HZ-31、JN654458 SDSU73、DQ988080 Ingelvac ATP、AY262352_HB-2(sh)2002、AY656990 Abst-1、AY032626 CH-1a、EU807840 CH-1R等经典株PRRSV的等位基因的核苷酸相似性为89.1%~95.8%;和JN654459 NADC30、KP860909 FJZ03、MH651746 SDQZ-1609、MH651747 SDZC-1609、KT945017 HNjz15、MH651748 TJZH-1607、KP861625 CHsx1401等NADC30类PRRSV的等位基因的核苷酸相似性为85.2%~87%。
ORF5的推导氨基酸有200个,碱性氨基酸18个,酸性氨基酸12个,疏水性氨基酸87个,极性氨基酸60个,等电点8.474。RRSV HN株的ORF5和EF112447 HEB1、MF370557 FZ06A、EF075945 HUB1、FJ548855 JXA1 P15、EU200962 Henan-1、EU624117 XH-GDDE等高致性PRRSV的等位基因的推导氨基酸相似性为98.4%~99.0% ;与KC445138 HZ-31、JN654458 SDSU73、DQ988080 Ingelvac ATP、AY262352_HB-2(sh)2002、AY656990 Abst-1、AY032626 CH-1a、EU807840 CH-1R等经典株PRRSV的等位基因的推导氨基酸相似性为86.8%~94.9%;与JN654459 NADC30、KP860909 FJZ03、MH651746 SDQZ-1609、MH651747 SDZC-1609、KT945017 HNjz15、MH651748 TJZH-1607、KP861625 CHsx1401等NADC30类PRRSV的等位基因的推导氨基酸相似性为84%~85.5%。表2,图2
PRRSV HNHK3-2021和EU624117 XH-GD的遗传距离最近,其与疫苗毒FJ548855 JXA1 P15在同一分支,在对该毒株的防控中可以对FJ548855 JXA1 P15对应的疫苗优先考虑,并且PRRSV HNHK3-2021与EF112447 HEB1、MF370557 FZ06A、EF075945 HUB1、EU200962 Henan-1等高致性PRRSV属于同一分支;与KC445138 HZ-31、JN654458 SDSU73、DQ988080 Ingelvac ATP、AY262352_HB-2(sh)2002、AY656990 Abst-1、AY032626 CH-1a、EU807840 CH-1R等经典株PRRSV的遗传距离较近与JN654459 NADC30、KP860909 FJZ03、MH651746 SDQZ-1609、MH651747 SDZC-1609、KT945017 HNjz15、MH651748 TJZH-1607、KP861625 CHsx1401等NADC30类PRRSV的遗传距离较远。
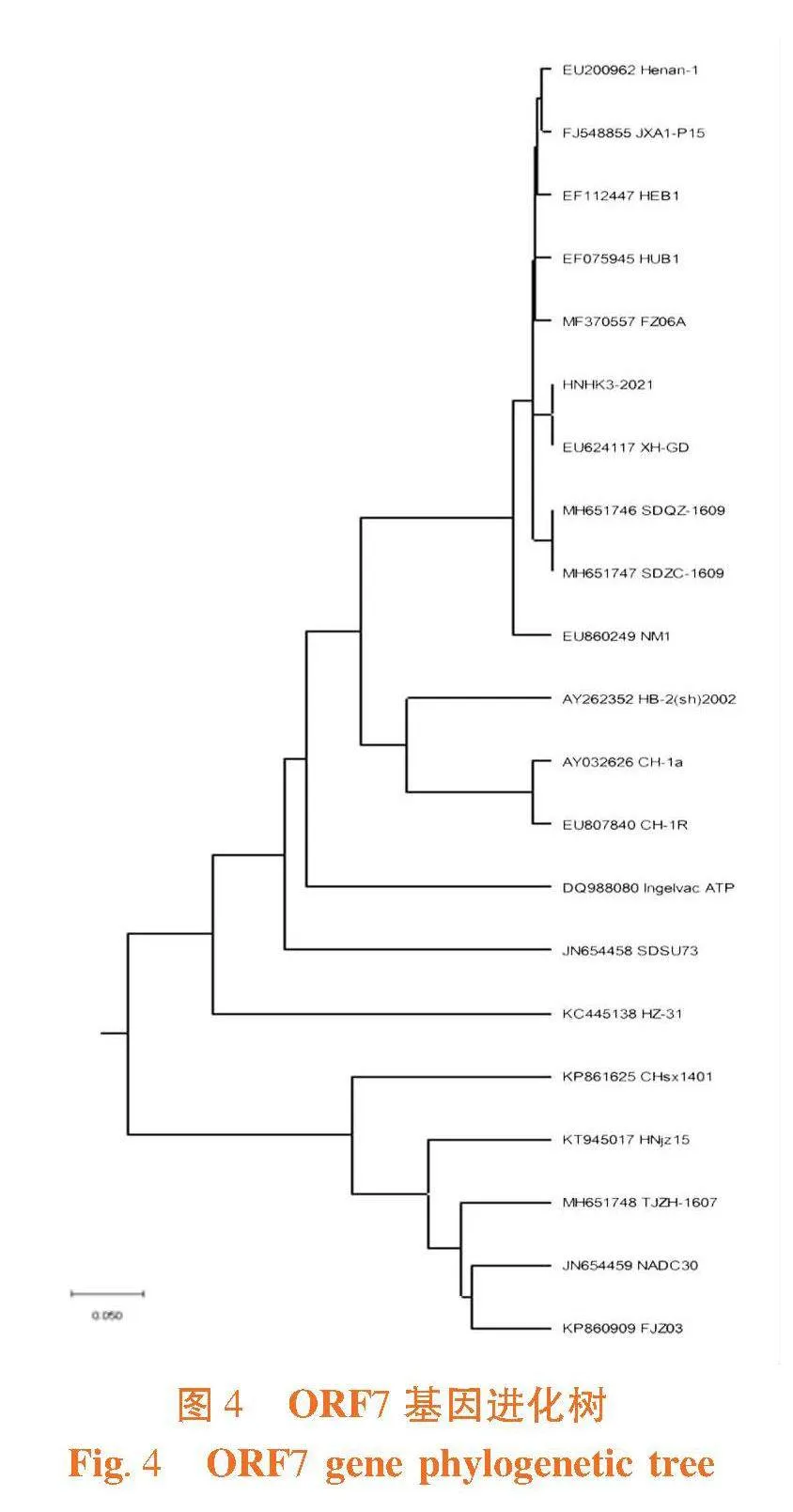
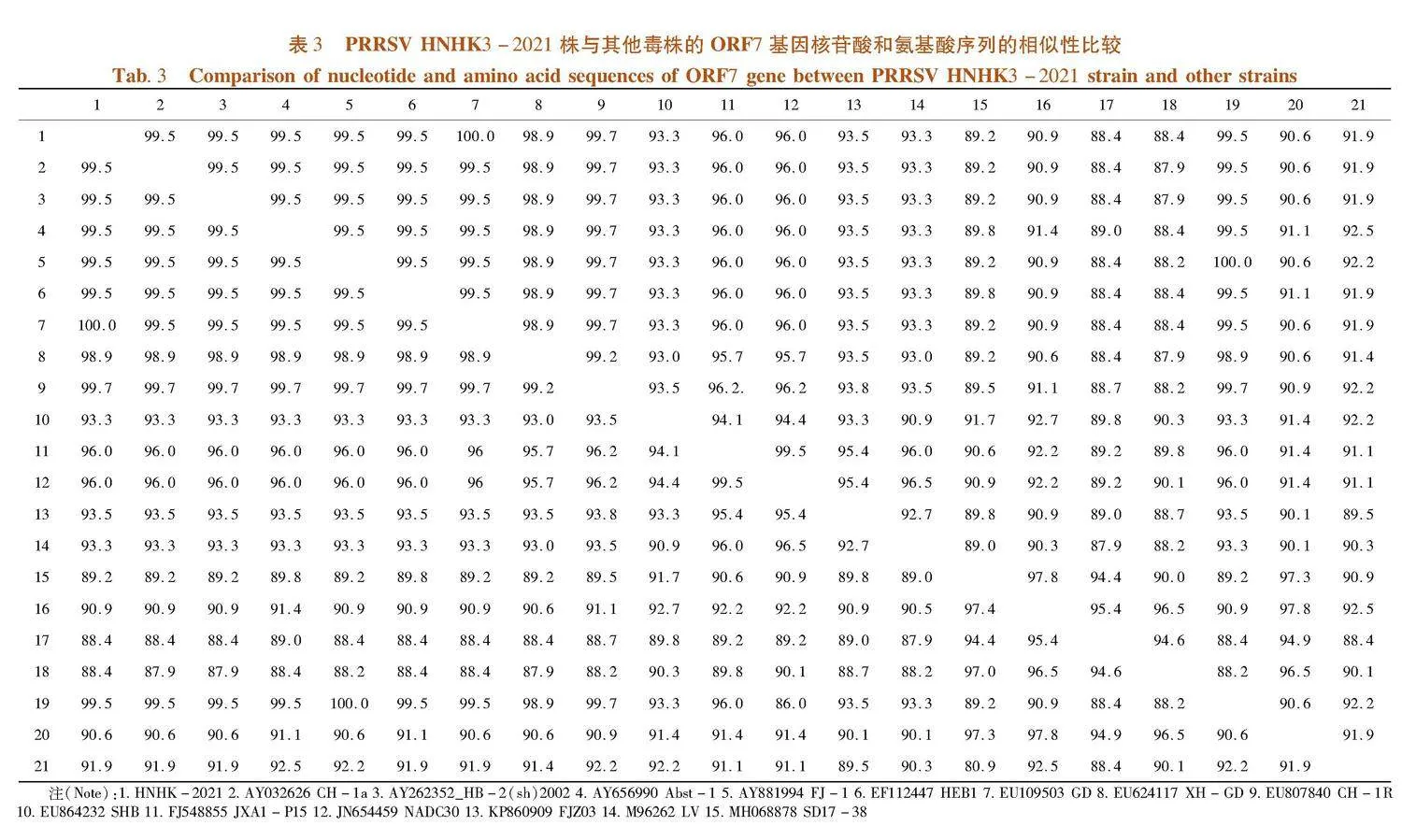
ORF7片段长度为372 bp,PRRSV HNHK3-2021株的ORF7与EF112447 HEB1、MF370557 FZ06A、EF075945 HUB1、FJ548855 JXA1 P15、EU200962 Henan-1、EU624117 XH-GDDE等高致性PRRSV的等位基因的核苷酸相似性为99.5%~100% ;与KC445138 HZ-31、JN654458 SDSU73、DQ988080 Ingelvac ATP、AY262352_HB-2(sh)2002、AY656990 Abst-1、AY032626 CH-1a、EU807840 CH-1R等经典株PRRSV的等位基因的核苷酸相似性为91.9%~96%;与JN654459 NADC30、KP860909 FJZ03、MH651746 SDQZ-1609、MH651747 SDZC-1609、KT945017 HNjz15、MH651748 TJZH-1607、KP861625 CHsx1401等NADC30类PRRSV的等位基因的核苷酸相似性为88.4%~89.2%。表3,图3
ORF7的推导氨基酸有123个,碱性氨基酸19个酸性氨基酸7个,疏水性氨基酸29个,极性氨基酸43个,等电点10.208。PRRSV HNHK3-2021株的ORF7与EF112447 HEB1、MF370557 FZ06A、EF075945 HUB1、FJ548855 JXA1 P15、EU200962 Henan-1、EU624117 XH-GDDE等高致性PRRSV的等位基因的推导氨基酸相似性为100% ;与KC445138 HZ-31、JN654458 SDSU73、DQ988080 Ingelvac ATP、AY262352_HB-2(sh)2002、AY656990 Abst-1、AY032626 CH-1a、EU807840 CH-1R等经典株PRRSV的等位基因的推导氨基酸相似性为92.7%~96%;与JN654459 NADC30、KP860909 FJZ03、MH651746 SDQZ-1609、MH651747 SDZC-1609、KT945017 HNjz15、MH651748 TJZH-1607、KP861625 CHsx1401等NADC30类PRRSV的等位基因的推导氨基酸相似性为88.7%~90.3%。
PRRSV HNHK3-2021和EU624117 XH-GD的遗传距离最近,其与疫苗毒FJ548855 JXA1 P15在同一分支,在对该毒株的防控中选取灭活疫苗JXA1,并且PRRSV HNHK3-2021与EF112447 HEB1、MF370557 FZ06A、EF075945 HUB1、EU200962 Henan-1、等高致性PRRSV属于同一分支;与KC445138 HZ-31、JN654458 SDSU73、DQ988080 Ingelvac ATP、AY262352_HB-2(sh)2002、AY656990 Abst-1、AY032626 CH-1a、EU807840 CH-1R等经典株PRRSV的遗传距离较近与JN654459 NADC30、KP860909 FJZ03、MH651746 SDQZ-1609、MH651747 SDZC-1609、KT945017 HNjz15、MH651748 TJZH-1607、KP861625 CHsx1401等NADC30类PRRSV的遗传距离较远。图4
3" 讨 论
3.1
将PRRSV HNHK3-2021与数据库中的不同毒株进行对比,其ORF5和ORF7片段都相较于EU624117 XH-GD、EF112447 HEB1、EU109503 GD更加接近。该病毒作为高致病性毒株,其NSP2缺失位点同结果与文献等[12-14]的研究结果一致。
我国在1995年出现PRRS病例,1996年报道分离出以 CH-1a为代表的经典型毒株[15]。2006年发现以高热、高发病、高死亡的高致病性毒株[16],其在NSP2中具有30个氨基酸的缺失[17],该毒株造成猪死亡[18]。在2013年我国又出现了PRRSV的流行,是由新的NADC30-like毒株所引起的。
3.2
GP5对于病毒复制具有重要意义[19]。ORF5基因是不同亚型的PRRSV存在差异最大的区域之一,欧洲型PRRSV毒株和美洲型PRRSV毒株在此区域只有51%~55%的序列相同,因此在大多数情况下ORF5可以起到病毒分型的作用。N蛋白由ORF7基因编码,为核衣壳蛋白,为病毒粒子的主要结构蛋白之一,N蛋白可以形成由二硫键所连接的同源二聚体,功能是包装病毒基因组RNA,并且N蛋白是唯一已知的不编码跨膜结构域或在PRRSV病毒粒子上无外结构域的结构蛋白。灭活疫苗不能保护猪免受高致病性PRRSV的攻击[20]。PRRSV HN在进化树上和疫苗毒FJ548855 JXA1 P15在同一分支,因此在今后对PRRSV HN的防治中可以优先使用疫苗 JXA1等高致病性疫苗株进行免疫。
4" 结 论
将海南分离到PRRSV HNHK3-2021株的GP5、GP7基因序列与GenBank上登录的21株PRRSVGP5、GP7基因序列构建遗传进化树显示,与EU624117 XH-GD(广东江门分离株)亲缘关系最近,为高致病性毒株,属于美洲型变异株。RRSV HNHK3-2021 毒株和EU624117 XH-GD具有最近的遗传距离。
参考文献(References)
[1]Wensvoort G, Terpstra C, Pol J M, et al. Mystery swine disease in The Netherlands: the isolation of Lelystad virus[J]. The Veterinary Quarterly, 1991, 13(3): 121-130.
[2] 周峰, 郭龙飞, 刘红英, 等. 猪繁殖与呼吸综合征病毒XINX株的分离及NSP2基因的遗传进化分析[J]. 江苏农业科学, 2014, 42(9): 180-182.
ZHOU Feng, GUO Longfei, LIU Hongying, et al. Isolation of porcine reproductive and respiratory syndrome virus XINX strain and genetic evolution analysis of NSP2 gene[J]. Jiangsu Agricultural Sciences," 2014, 42(9): 180-182.
[3] 王亚磊, 茆达干, 孟春花, 等. PRRSV非结构蛋白与病毒逃逸宿主免疫反应的机制[J]. 江苏农业科学, 2014, 42(5): 165-167.
WANG Yalei, MAO Dagan, MENG Chunhua, et al. Mechanism of non-structural protein of PRRSV and virus escaping from host immune response[J]. Jiangsu Agricultural Sciences," 2014, 42(5): 165-167.
[4] Allaire M, Chernaia M M, Malcolm B A, et al. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases[J]. Nature," 1994, 369(6475): 72-76.
[5] Zhou S C, Ge X N, Kong C, et al. Characterizing the PRRSV nsp2 deubiquitinase reveals dispensability of Cis-activity for replication and a link of nsp2 to inflammation induction[J]. Viruses, 2019, 11(10): 896.
[6] Weiland E, Wieczorek-Krohmer M, Kohl D, et al. Monoclonal antibodies to the GP5 of porcine reproductive and respiratory syndrome virus are more effective in virus neutralization than monoclonal antibodies to the GP4[J]. Veterinary Microbiology, 1999, 66(3): 171-186.
[7] Guzmán M, Meléndez R, Jiménez C, et al. Analysis of ORF5 sequences of Porcine Reproductive and Respiratory Syndrome virus (PRRSV) circulating within swine farms in Costa Rica[J]. BMC Veterinary Research," 2021, 17(1): 217.
[8] Olasz F, Dénes B, Bálint, et al. Immunological and biochemical characterisation of 7ap, a short protein translated from an alternative frame of ORF7 of PRRSV[J]. Acta Veterinaria Hungarica, 2016, 64(2): 273-287.
[9] Bálint, Molnár T, Kecskeméti S, et al. Genetic variability of PRRSV vaccine strains used in the national eradication programme, Hungary[J]. Vaccines," 2021, 9(8): 849.
[10] 贺冬梅, 谭树义, 陈朝喜, 等. PRRSV HuN-2011株ORF5和ORF7基因的序列分析[J]. 黑龙江畜牧兽医, 2017(7): 188-191.
HE Dongmei, TAN Shuyi, CHEN Zhao Xi, et al. Sequence analysis of ORF5 and ORF7 genes of PRRSV HuN-2011 strain[J]. Heilongjiang Animal Science and Veterinary Medicine," 2017(7): 188-191.
[11] 秦宏阳, 曹宗喜, 汤国凯, 等. 4株PRRSV GP5蛋白编码序列的序列测定和特性分析[J]. 广东畜牧兽医科技, 2008, 33(5): 25-28.
QIN Hongyang, CAO Zongxi, TANG Guokai, et al. Sequencing and analysis of four PRRSV strains GP5 proteins[J]. Guangdong Journal of Animal and Veterinary Science, 2008, 33(5): 25-28.
[12] Cao Z X, Jiao P R, Huang Y M, et al. Genetic diversity analysis of the ORF5 gene in porcine reproductive and respiratory syndrome virus samples from South China[J]. Acta Veterinaria Hungarica," 2012, 60(1): 157-164.
[13] 曹宗喜, 潘全会, 林哲敏, 等. PRRSV广东株XH-GD的分离及其NSP2、ORF3、ORF5的序列分析[J]. 广东农业科学, 2013, 40(12): 159-163.
CAO Zongxi, PAN Quanhui, LIN Zhemin, et al. Isolation and sequence analysis of NSP2, ORF3 and ORF5 of PRRSV XH-GD strain from Guangdong Province[J]. Guangdong Agricultural Sciences, 2013, 40(12): 159-163.
[14] Guo Z H, Chen X X, Li R, et al. The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective[J]. Virology Journal," 2018, 15(1): 2.
[15] 郭宝清, 陈章水. 哈兽研所首次证实国内猪群存在PRRSV感染[J]. 兽医科技信息, 1996,(1): 8.
GUO Baoqing, CHEN Zhangshui. Harbin Institute of Veterinary Medicine confirmed for the first time that PRRSV infection exists in domestic pigs[J]. Chinese Journal of Animal Husbandry and Veterinary Medicine," 1996,(1): 8.
[16] 吴鹏, 张勋, 梁田, 等. 北疆地区猪繁殖与呼吸综合征病毒ORF7基因克隆与遗传演化分析[J]. 新疆农业科学, 2016, 53(11): 2149-2156.
WU Peng, ZHANG Xun, LIANG Tian, et al. PRRSV ORF7 cloning and genetic evolution analysis of pigs in northern Xinjiang[J]. Xinjiang Agricultural Sciences," 2016, 53(11): 2149-2156.
[17] An T Q, Zhou Y J, Liu G Q, et al. Genetic diversity and phylogenetic analysis of glycoprotein 5 of PRRSV isolates in mainland China from 1996 to 2006: coexistence of two NA-subgenotypes with great diversity[J]. Veterinary Microbiology, 2007, 123(1/2/3): 43-52.
[18] 陆桂丽, 汪萍, 成进, 等. 猪繁殖与呼吸综合征新疆株ORF2-7基因的克隆及序列分析[J]. 新疆农业科学, 2014, 51(3): 546-551.
LU Guili, WANG Ping, CHENG Jin, et al. Clone and sequence analysis of ORF2-7 genes of porcine reproductive and respiratory syndrome virus of Xinjiang strains[J]. Xinjiang Agricultural Sciences, 2014, 51(3): 546-551.
[19] Suárez P, Zardoya R, Martín M J, et al. Phylogenetic relationships of European strains of porcine reproductive and respiratory syndrome virus (PRRSV) inferred from DNA sequences of putative ORF-5 and ORF-7 genes[J]. Virus Research, 1996, 42(1/2): 159-165.
[20]王旭哲, 刘永宏, 赵丽, 等. 新疆南疆猪繁殖与呼吸综合征病毒ORF5基因克隆与序列分析[J]. 新疆农业科学, 2013, 50(2): 357-364.
WANG Xuzhe, LIU Yonghong, ZHAO Li, et al. The Cloning and the Sequence Analysis of Southern Xinjiang Strain ORF5 Gene of Porcine Reproductive and Respiratory Syndrome Virus[J]. Xinjiang Agricultural Sciences," 2013, 50(2): 357-364.
Sequence analysis of ORF3 and ORF5 of PRRSV HNHK3-2021 strain isolated in Hainan
FAN Yuexuan1,2, MA Jiamei1,2, HUANG Chunyuan2,ZHENG Jiaxin2, CHEN Suzhen1,2, ZHANG Yan2, LIU Guangliang1,3,CAO Zongxi1,2
(1. College of Veterinary Medicine, Xinjiang Agricultural University, Urumqi 830052, China; 2. Hainan Provincial Key Laboratory of Tropical Animal Breeding and Disease Research, Institute of Animal Husbandry and Veterinary Medicine, Hainan Academy of Agricultural Sciences, Haikou 571100, China; 3. Hainan Provincial Academician Innovation Center, Institute of Animal Husbandry and Veterinary Medicine, Hainan Academy of Agricultural Sciences, Haikou 571100, China)
Abstract:【Objective】 To explore the epidemiological characteristics and genetic variation of PRRSV in Hainan.
【Methods】" Samples were collected from Haikou and surrounding areas.The PRRSV strain HNHK3-2021 was isolated, and the NSP2 genes, as well as the ORF5 and ORF7 gene sequences, were determined.Sequence alignment analysis was conducted using Lasergene 7.1, and genetic evolution analysis was performed using MEGA-X.
【Results】" The PRRSV HNHK3-2021 strain isolated was found to be of American origin with high nucleotide similarity to EF112447 HEB1, EU109503 GD, and EU624117 XH-GD ORF5 and ORF7 (99.2%-99.3% and 99.5%-100%, respectively).The derived amino acid similarity was 99%-99.5% and 100%.The nucleotide similarities of ORF5 and ORF7 of PRRSV HNHK3-2021 strain and EU864232 SHB, AY262352_HB-2(sh)2002, AY656990 Abst-1, AY032626 CH-1a, EU807840 CH-1R were found to be 87.1% -96.4% and 91.4%-97.3%, respectively.Similarly, the similarity of amino acids was 86.6%-93% and 91.9%-97.6%.The nucleotide similarities of ORF5 and ORF7 of PRRSV HNHK3-2021 strain and JN654459 NADC30, KP860909 FJZ03, MH068878 SD17-38, AY881994 FJ-1 were found to be 85.4%-87.1% and 89%-91.4%.The similarity of amino acids was 86.1%-87.1% and 89.5%-91.9%.
【Conclusion】 The PRRSV HNHK3-2021 strain and EU624117 XH-GD have the closest genetic distance.
Key words:porcine reproductive and respiratory syndrome; genetic evolution analysis; ORF5; ORF7
Fund projects:Technological Innovation Project of Hainan Provincial Scientific Research Institutes(jscx202001);The National Natural Science Foundation of China(32060796);Open Fund of the State Key Laboratory of Pathogenic Biology of Livestock Diseases(SKLVEB2020KFKT021);Hainan Provincial Academy of Agricultural Sciences Research Project at the same level(HAAS2022PT0205);National Key Ramp;D Program of China(2023YFC3404302)
Correspondence author:CAO Zongxi (1982- ), male, from Xinxiang Henan, researcher, doctor, doctoral supervisor, research direction: livestock infectious diseases, (E-mail)caozongxi@163.com
LIU Guangliang (1975- ), male, from Zizhong Sichuan, researcher, doctor, doctoral supervisor, research direction:animal vaccine development and molecular immunology, (E-mail)liuguangliang01@caas.cn